Using the Heatmap
There are four heatmap states that can be used on the molecular data page: Ranked Genes, Clustered Expression, Marker Genes and Custom.
- Ranked genes: displays the top genes according to DE calculations for a cell label or cluster. See Running Differential Expression for more information.
- Clustered Expression: displays the top genes clustered hierarchically by expression pattern for a selected set of cells. This can be used to analyze gene expression in broad clusters in order to break them in to subclusters.
- Marker Genes: displays a heatmap with the marker genes provided by the scientist(s) for the dataset.
- Custom: displays a custom set of genes selected by the user.
NOTES:
- The heatmap displays the top three genes from the top ten cell labels by number of cells. All of the cell labels are displayed on the horizontal axis of the heatmap but the number of genes displayed on the vertical axis is limited to 30.
- The top genes in the heatmap and gene table are sorted by log2FC.
Expanding and collapsing the heatmap view:
- The heatmap can be expanded by clicking the Enlarge Heatmap icon in the top right corner of the heatmap and the expanded view can be exited by clicking the same icon to return to the regular view.
Expanding the heatmap for a selected cluster:
- Select cluster or cell labels on the embedding or in the Sets of Cell Labels in the left panel. The selected cells will be colored red on the embedding and in the bar above the heatmap. The view of the selected cells can be expanded by clicking on the Expand Selection icon.
Using the Clustered Expression heatmap:
- Select the cluster or cell label on the embedding or by selecting the cell label in the side bar. Once the cells have been selected, click on Clustered Expression on the heatmap to open the clustered expression heatmap.
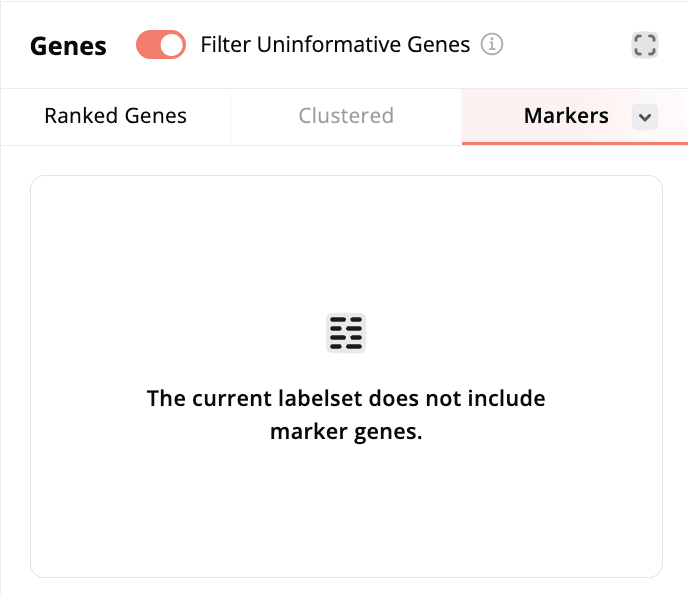
Using the Marker Genes heatmap:
- To use the marker genes heatmap, click on More above the heatmap and then select Marker Genes. The heatmap will display the marker genes provided by the scientist(s) for each cell label with the corresponding cell label denoted by the color-coded bar on the edge of the heatmap.
- If the dataset does not contain marker genes, clicking on Marker Genes will display a message that the labelset does not contain marker genes.
Using the Custom heatmap:
- To use the custom heatmap, click on Custom. Next, select genes to add to the heatmap by clicking on the empty box next to the gene in the gene table or search for a specific gene of interest. Genes can be removed from the heatmap by clicking the checkmark next to the gene in the heatmap or by resetting all of the selected genes.
- You can also create a custom heatmap using the marker genes for a cell label by navigating to the metadata summary for the cell label in the side bar and clicking on Show on the heatmap in the marker gene section.
